Advanced Biotech Miscellaneous
- Choose the appropriate pair of primers to amplify the following DNA fragment by the polymerase chain reaction (PCR).
5′ –gacctgtgg------------------atacggGat –3′
3′ –ctggacacc-------------------tatgcccta –5′
Primers
P. 5′ –gacctgtgg–3′
Q. 5′ –ccacaggtc–3′
R. 5′ –tagggcata–3′
S. 5′ –atcccgtat–3′
-
View Hint View Answer Discuss in Forum
Respective primers will be anneal to 3′ ends of the two template strands; keeping antiparallel nature of DNA duplex and complimentarity in the mind.
Correct Option: B
Respective primers will be anneal to 3′ ends of the two template strands; keeping antiparallel nature of DNA duplex and complimentarity in the mind.
- A linear double stranded DNA of length 8 kbp has three restriction sites. Each of these can either be a BamHi or a HaeIII site. The DNA was digested completely with both enzymes. The products were purified and subjected to an end-filling reaction using the Klenow fragment and [α– 32P]-dCTP. The products of the end-filling reaction were purified, resolved by electrophoresis, stained with ethidium bromide (EtBr) and then subjected to autoradiography. The corresponding images are shown below.
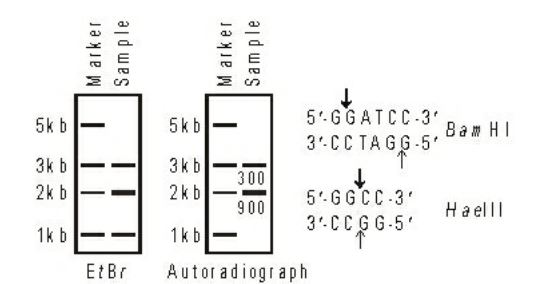
The numbers below each band in the sample lane in the autoradiograph represent their mean signal intensity in arbitrary units. Which one of the following options is the correct restriction map of the DNA ?
-
View Hint View Answer Discuss in Forum
Hae III : GG|CC
CC | GG
BamHI : G | GATC
CCTAG | G Correct Option: A
Hae III : GG|CC
CC | GG
BamHI : G | GATC
CCTAG | G
- Assuming random distribution of nucleotides, the average number of fragments generated upon digestion of a circular DNA of size 4.3 × 105 bp with AluI(5′-AG↓CT-3′) is ______ × 103.
-
View Hint View Answer Discuss in Forum
Average number of fragments generated = 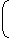
1 × 1 × 1 × 1 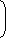
× 4.3 × 105 4 4 4 4 = 4.3 × 105 = 1.68 × 103 256 Correct Option: B
Average number of fragments generated = 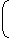
1 × 1 × 1 × 1 
× 4.3 × 105 4 4 4 4 = 4.3 × 105 = 1.68 × 103 256
- Plasmid DNA (0.5 µg) containing an ampicillin resistance marker was added to 200 µl of competent cells. The transformed competent cells were diluted 10,000 times, out of which 50 µl was plated on agar plates containing ampicillin. A total of 35 colonies were obtained. The transformation efficiency is _____ × 106 cfu. µg– 1.
-
View Hint View Answer Discuss in Forum
DNA= 0.5 µg
Dillution = 50 = 2.5 × 10-5 200 × 10000 TE = Number of colonies Dilution × Amount of DNA = 35 = 2.8 × 106 2.5 × 10-5 × 0.5 Correct Option: C
DNA= 0.5 µg
Dillution = 50 = 2.5 × 10-5 200 × 10000 TE = Number of colonies Dilution × Amount of DNA = 35 = 2.8 × 106 2.5 × 10-5 × 0.5
- The plasmid DNA was subjected to restriction digestion using the enzyme EcoRI and analysed on an agarose gel. Assuming digestion has worked (the enzyme was active), match the identity of the DNA bands shown in the image in Group I with their identity in Group II.
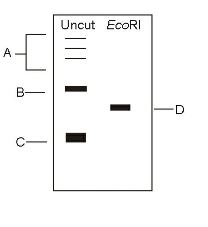
Group–I Group–II P. Bands labeled as A 1. Nicked Q. Band labeled as B 2. Supercoiled R. Band labeled as C 3. Concatemers S. Band labeled as D 4. Linear
-
View Hint View Answer Discuss in Forum
The bands which are labeled as A were uncut and separated at three different regions on the agarose gel after digestion with EcoRI. It shows that the plasmid A separates is having concatemers. Plasmid B labeled as B is nicked while Plasmid C which is labeled as C has a very thick band which suggests that Plasmid C is supercoiled. Band D was the only plasmid which was the digested product of Eco RI are linear because EcoRI has already digested the plasmid.
Correct Option: A
The bands which are labeled as A were uncut and separated at three different regions on the agarose gel after digestion with EcoRI. It shows that the plasmid A separates is having concatemers. Plasmid B labeled as B is nicked while Plasmid C which is labeled as C has a very thick band which suggests that Plasmid C is supercoiled. Band D was the only plasmid which was the digested product of Eco RI are linear because EcoRI has already digested the plasmid.

