Advanced Biotech Miscellaneous
- Identify the CORRECT statements
P. 5′ and 3′ ends of the transcripts can be mapped by utilizing polymerase chain reaction
Q. S1 nuclease can cleave the DNA strand of a DNA-RNA hybrid
R. T4 polynucleotide kinase is used for labeling 3′ end of DNA
S. Baculovirus (Autographa californica) can be used as an insect expression vector
-
View Hint View Answer Discuss in Forum
Rapid amplification of cDNA ends (RACE) polymerase chain reactions (PCR) are used for independent high-throughput verification of transcriptional starting sites (TSSs) determined by genome-wide assays. 52 -RACE PCR is a well-established and widely used method to specifically amplify the 52 end of a transcript, facilitating mapping of the TSS and the approximate location of promoter elements. Conventionally, this mapping is done by cloning the 52 -RACE PCR product into a bacterial vector and sequencing a few clones by classic electrophoresis-based Sanger sequencing. Now-a-days, even 3’ end transcripts are being mapped using 3’-RACE PCR. Baculoviruses are a very diverse group of viruses with double-stranded, circular, supercoiled genomes, with sizes varying from about 80 to over 180 kb which encode between 90 and 180 genes. Baculoviruses have evolved to initiate infection in the insect midgut. Baculovirus-insect cell expression systems have the capacity to produce many recombinant proteins at high levels and they also provide significant eukaryotic protein processing capabilities.
Correct Option: C
Rapid amplification of cDNA ends (RACE) polymerase chain reactions (PCR) are used for independent high-throughput verification of transcriptional starting sites (TSSs) determined by genome-wide assays. 52 -RACE PCR is a well-established and widely used method to specifically amplify the 52 end of a transcript, facilitating mapping of the TSS and the approximate location of promoter elements. Conventionally, this mapping is done by cloning the 52 -RACE PCR product into a bacterial vector and sequencing a few clones by classic electrophoresis-based Sanger sequencing. Now-a-days, even 3’ end transcripts are being mapped using 3’-RACE PCR. Baculoviruses are a very diverse group of viruses with double-stranded, circular, supercoiled genomes, with sizes varying from about 80 to over 180 kb which encode between 90 and 180 genes. Baculoviruses have evolved to initiate infection in the insect midgut. Baculovirus-insect cell expression systems have the capacity to produce many recombinant proteins at high levels and they also provide significant eukaryotic protein processing capabilities.
- Protein-protein interactions are studied by
P. DNA foot printing
Q. Yeast two hybrid system
R. Ligase chain reaction
S. Mass spectrometry
-
View Hint View Answer Discuss in Forum
Yeast two hybrid screening is a molecular biology technique used to discover protein-protein interaction and protein-DNA interaction by testing for physical interactions. Mass spectrometry (MS) is an analytical technique that measures the mass-to-charge ratio of charged particles.
Correct Option: B
Yeast two hybrid screening is a molecular biology technique used to discover protein-protein interaction and protein-DNA interaction by testing for physical interactions. Mass spectrometry (MS) is an analytical technique that measures the mass-to-charge ratio of charged particles.
- The study of evolutionary relationships is known as
-
View Hint View Answer Discuss in Forum
Phylogenetics deals with evolutionary relatedness of the species. It calculates the distances amongst the species from its origin and can track down any ancestral relationship.
Correct Option: C
Phylogenetics deals with evolutionary relatedness of the species. It calculates the distances amongst the species from its origin and can track down any ancestral relationship.
- A linear double stranded DNA of length 8 kbp has three restriction sites. Each of these can either be a BamHi or a HaeIII site. The DNA was digested completely with both enzymes. The products were purified and subjected to an end-filling reaction using the Klenow fragment and [α– 32P]-dCTP. The products of the end-filling reaction were purified, resolved by electrophoresis, stained with ethidium bromide (EtBr) and then subjected to autoradiography. The corresponding images are shown below.
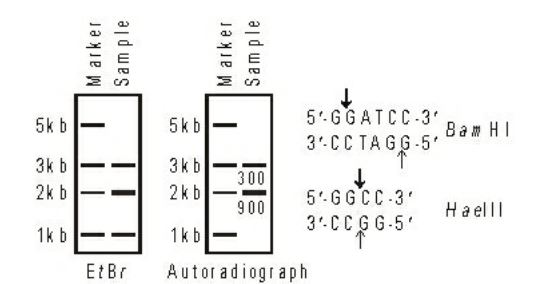
The numbers below each band in the sample lane in the autoradiograph represent their mean signal intensity in arbitrary units. Which one of the following options is the correct restriction map of the DNA ?
-
View Hint View Answer Discuss in Forum
Hae III : GG|CC
CC | GG
BamHI : G | GATC
CCTAG | G Correct Option: A
Hae III : GG|CC
CC | GG
BamHI : G | GATC
CCTAG | G
- Plasmid DNA (0.5 µg) containing an ampicillin resistance marker was added to 200 µl of competent cells. The transformed competent cells were diluted 10,000 times, out of which 50 µl was plated on agar plates containing ampicillin. A total of 35 colonies were obtained. The transformation efficiency is _____ × 106 cfu. µg– 1.
-
View Hint View Answer Discuss in Forum
DNA= 0.5 µg
Dillution = 50 = 2.5 × 10-5 200 × 10000 TE = Number of colonies Dilution × Amount of DNA = 35 = 2.8 × 106 2.5 × 10-5 × 0.5 Correct Option: C
DNA= 0.5 µg
Dillution = 50 = 2.5 × 10-5 200 × 10000 TE = Number of colonies Dilution × Amount of DNA = 35 = 2.8 × 106 2.5 × 10-5 × 0.5

